-
Notifications
You must be signed in to change notification settings - Fork 63
Pre Process
Some genes are expressed at extremely low levels in all samples. We need to remove these genes from further analysis. By default, a gene has to have more than 10 counts in at least one sample. Otherwise, the gene is removed.
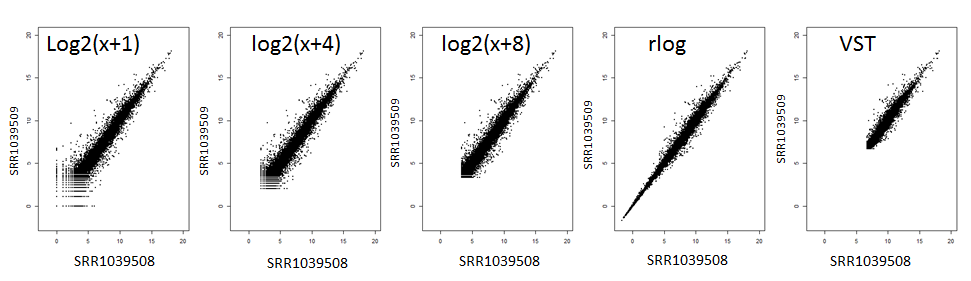
There are 3 options for transformation of counts data for clustering analysis and principal component analysis:
- VST: variance stabilizing transform
- rlog: regularized log (only for N<10)
- Sarted log: log2(x+c)
VST is performed according to (Anders and Huber 2010) and rlog is based on (Love, Huber et al. 2014). When there are more than 10 samples, rlog becomes slow. The default is started log, where a pseudo count c is added to all counts before log transformation. The constant c can range from 1 to 10. The bigger this number is, the less sensitive it is to noise from low counts. The effect of these transformations can be visualized below between technical replicates. You can see that VST is very aggressive in transforming the data.
For counts data, transformed data is only used for clustering analysis and PCA, MDS plots. Thus your choice does not affect the identification of differentially expressed genes (DEGs), fold-change, and pathway analysis, which are based on original counts data.
iDEP produces a barplot representing total read counts per library. When the library sizes are more than 3 times different, limma-trend method is not recommended for identifying DEGs. See the manual for limma.
For normalized expression data, a filter is also applied to remove genes expressed at low levels across all samples. By default, only genes expressed at the level of 1 or higher in at least one sample will be included for further analysis. This number works for Affymetrix microarrays, but it should be changed according to the data format. For cDNA microarrays, where the expression levels are log ratios, we need to set this to a large negative number such as -1e20 to disable this filter.
Users can choose to perform log transformation. iDEP calculates kurtosis for each of the data columns, and if the mean kurtosis is bigger than 50, a log2 transformation is enforced. Large kurtosis usually indicates the presence of extremely large numbers in the data set that warrants log-transformation.
Users can double check the effects of data transformation by examining the box plot and density plot on this page.
- Anders, S. and W. Huber (2010). “Differential expression analysis for sequence count data.” Genome Biol 11(10): R106.
- Love, M. I., W. Huber and S. Anders (2014). “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.” Genome Biol 15(12): 550.