-
Notifications
You must be signed in to change notification settings - Fork 6
Home 1_3
PanPhlAn is a strain-level metagenomic profiling tool for identifying the gene composition and in-vivo transcriptional activity of individual strains in metagenomic samples. PanPhlAn’s ability for strain-tracking and functional analysis of unknown pathogens makes it an efficient tool for culture-free infectious outbreak epidemiology and microbial population studies.
For a detailed tutorial with a small example dataset, see Tutorial. For help, use the bioBakery help forum.
The PanPhlAn software team: Léonard Dubois, Matthias Scholz (algorithm design), Moreno Zolfo (Roary import), Thomas Tolio (programmer), and Nicola Segata (principal investigator).
Matthias Scholz*, Doyle V. Ward*, Edoardo Pasolli*, Thomas Tolio, Moreno Zolfo, Francesco Asnicar, Duy Tin Truong, Adrian Tett, Ardythe L. Morrow, and Nicola Segata (* Equal contribution) Strain-level microbial epidemiology and population genomics from shotgun metagenomics Nature Methods, 13, 435–438, 2016.
PanPhlAn can be easily installed using pip
pip install panphlan==1.3
Otherwise, for a custom installation, one can retrieve the source code by cloning the GitHub repository
git clone https://github.com/SegataLab/panphlan.git
PanPhlAn runs under Ubuntu/Linux and requires the following software tools to be installed on your system:
Add the paths of your installed tools to your .bashrc file
export PATH=/your/path/to/samtools/:$PATH
export PATH=/your/path/to/bowtie2/bowtie2-2.1.0/:$PATH
export BT2_HOME=/your/path/to/bowtie2/bowtie2-2.1.0/
export BOWTIE2_INDEXES=/your/path/to/PanPhlAn_DB/and/bowtie2_indexes/
-
Pangenome generation
Create bowtie2 indexes for a bacterial species using references genomes and pangenome file (ChocoPhlAn export)
./panphlan_new_pangenome_generation.py -c speciesname --i_fna genome-files/ -o database/ -
PanPhlAn mapping
Map each metagenomic sample against the species database. Example: screen for E. coli pangenome genes in sample01 and sample02 (ecoli16 database)
./panphlan_map.py -c ecoli16 -i Samples/sample01.fastq -o map_results/sample01_ecoli16.csv
./panphlan_map.py -c ecoli16 -i Samples/sample02.fastq -o map_results/sample02_ecoli16.csv -
PanPhlAn profiling
Merge and process the mapping results for getting the final gene-family presence/absence profile matrix
./panphlan_profile.py -c ecoli16 -i map_results/ --o_dna result_gene_presence_absence.csv
Based on shotgun metagenomic samples, PanPhlAn enables:
- strain identification and characterization of unknown strains in metagenomic samples. The gene set of strains present in samples is detected by screening for all potential genes from the species pangenome.
- outbreak monitoring: pathogen detection and characterization, see E. coli example below.
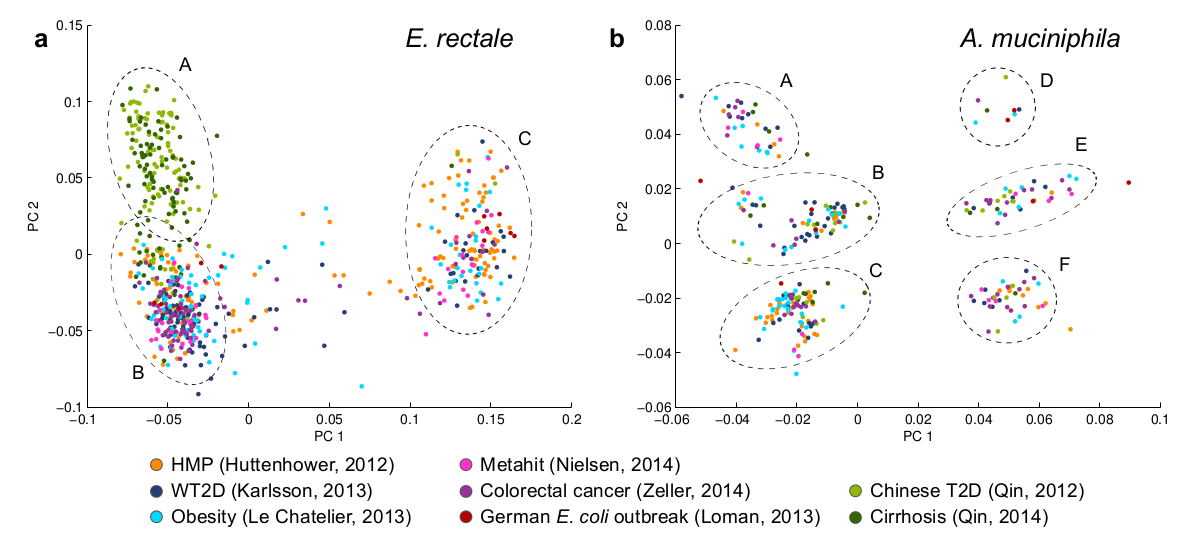
- population genomics: exploring the diversity of a species based on detected strains in hundreds of samples, see E. rectale and A. muciniphila examples below.
- strain tracking: detecting identical gene content profiles of strains in different samples
- functional analysis: based on detected strain-specific genes, the gene sequences can be used for functional investigations using KEGG or other similar databases.
- in-vivo transcriptional activity: with focus on genes that are specific to the individual strain in a sample (species and strain specific transcriptomics).
PanPhlAn profiling of the German outbreak metagenomes using a reference database in which the target outbreak genome is missing. (a) Hierarchical clustering. The heatmap displays presence/absence gene-family profiles of 110 reference strains (bright colored columns) and of 12 metagenomically detected strains (darker columns). Most outbreak samples cluster together due to almost identical profiles (right), with four samples (left) showing different profiles due to the presence of additional dominant E. coli strains overlying the target outbreak strain. (b) Functional analysis of outbreak-specific gene-families (Fisher exact test) confirmed that the outbreak strain is a combination of a EAEC pathogen (pAA plasmid) with acquired Shiga toxin and antibiotic resistance genes, complemented with a set of enriched virulence-related functions and pathway modules.
Large-scale population genomics study of E. rectale and A. muciniphila. Based on 1830 metagenomic samples from 8 cohorts, PanPhlAn reveals the subspecies structure even when only few species reference genomes are available. (a) Based on only one reference genome, E. rectale strains can be resolved into three geographically distinct clades. Clade A is related to samples of the two Chinese cohorts (bright and dark green dots). (b) Based on two available reference genomes, PanPhlAn shows a clear cluster structure of A. muciniphila strains, suggesting that the species can be distinguished into six functionally distinct clades A-F.
- Designed to work with Python 3; Python 2.7 compatibility is not guaranteed.
- Major changes in
panphlan_pangenome_generation.py. It now works using ChocoPhlan and only generates bowtie2 indexes -
panphlan_old_pangenome_generation.pyworks as in the previous version. For custom database generation using usearch -
panphlan_profile.pyadd arguments to map genes ID to functionnal annotation data
-
panphlan_pangenome_generation.pyRoary clustering is accepted as input for database generation (instead of usearch7 clustering)
-
panphlan_pangenome_generation.pygff gene-annotation files are accepted as input for database generation (instead ffn gene sequence input)
- Stable release published together with the PanPhlAn paper in Nature Methods (March 2016)
-
panphlan_pangenome_generation.pygene-names are no more restricted to NCBI coding -
panphlan_profile.pyset 2X as default min coverage threshold of a strain in sample
- First release of PanPhlAn as used for the analysis in the PanPhlAn paper
PanPhlAn is a project of the Computational Metagenomics Lab at CIBIO, University of Trento, Italy.
- PanPhlAn 3.0
- PanPhlAn 1.3