diff --git a/README.md b/README.md
index 2cbb2e92..ab0cbe57 100644
--- a/README.md
+++ b/README.md
@@ -32,13 +32,15 @@ Latest Update: follow up by clicking `Starred` and `Watch` on our [GitHub repos
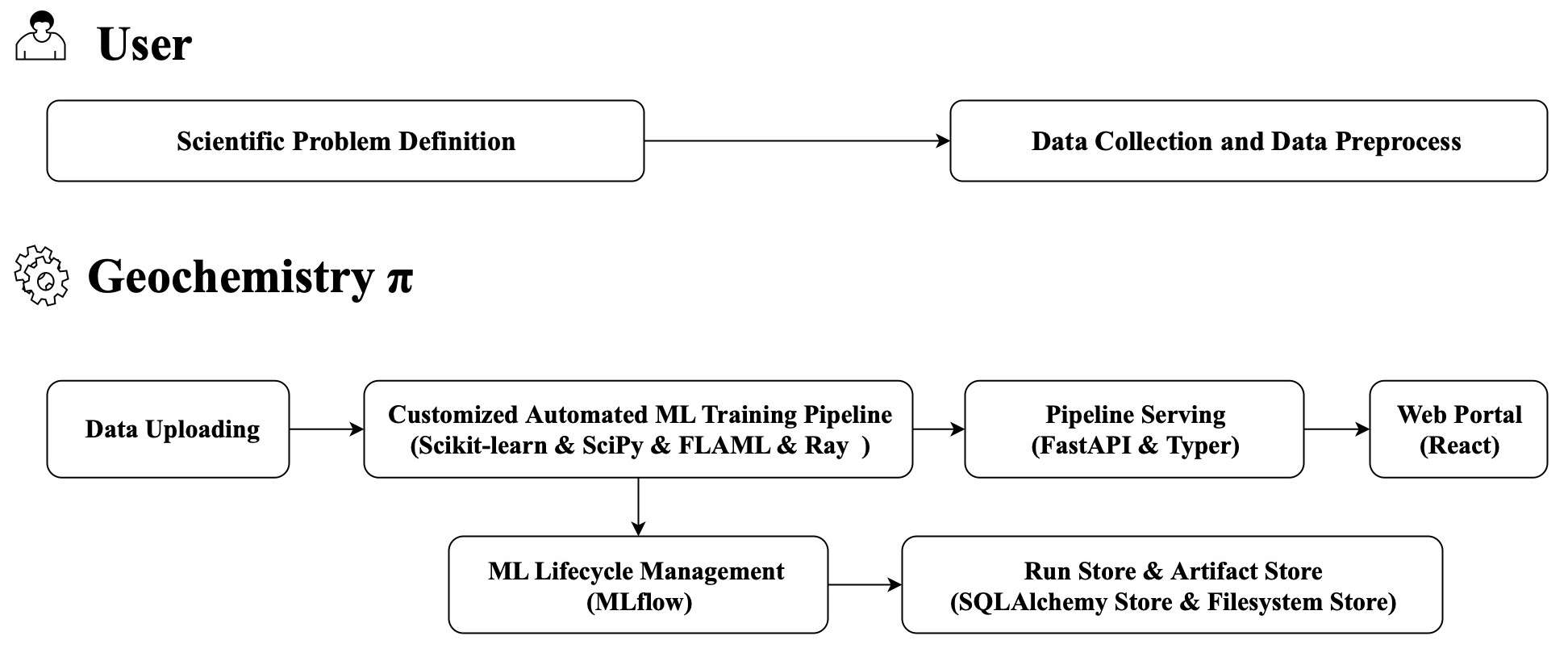
The following figure is the simplified overview of Geochemistry π:
-
+
+  +
+
The following figure is the frontend-backend separation architecture of Geochemistry:
-
-

-
+  +
+
## Quick Installation
@@ -140,6 +142,11 @@ Its data section provides feature engineering based on **arithmatic operation**.
Its models section provides both **supervised learning** and **unsupervised learning** methods from **Scikit-learn** framework, including four types of algorithms, regression, classification, clustering, and dimensional reduction. Integrated with **FLAML** and **Ray** framework, it allows the users to run AutoML easily, fastly and cost-effectively on the built-in supervised learning algorithms in our framework.
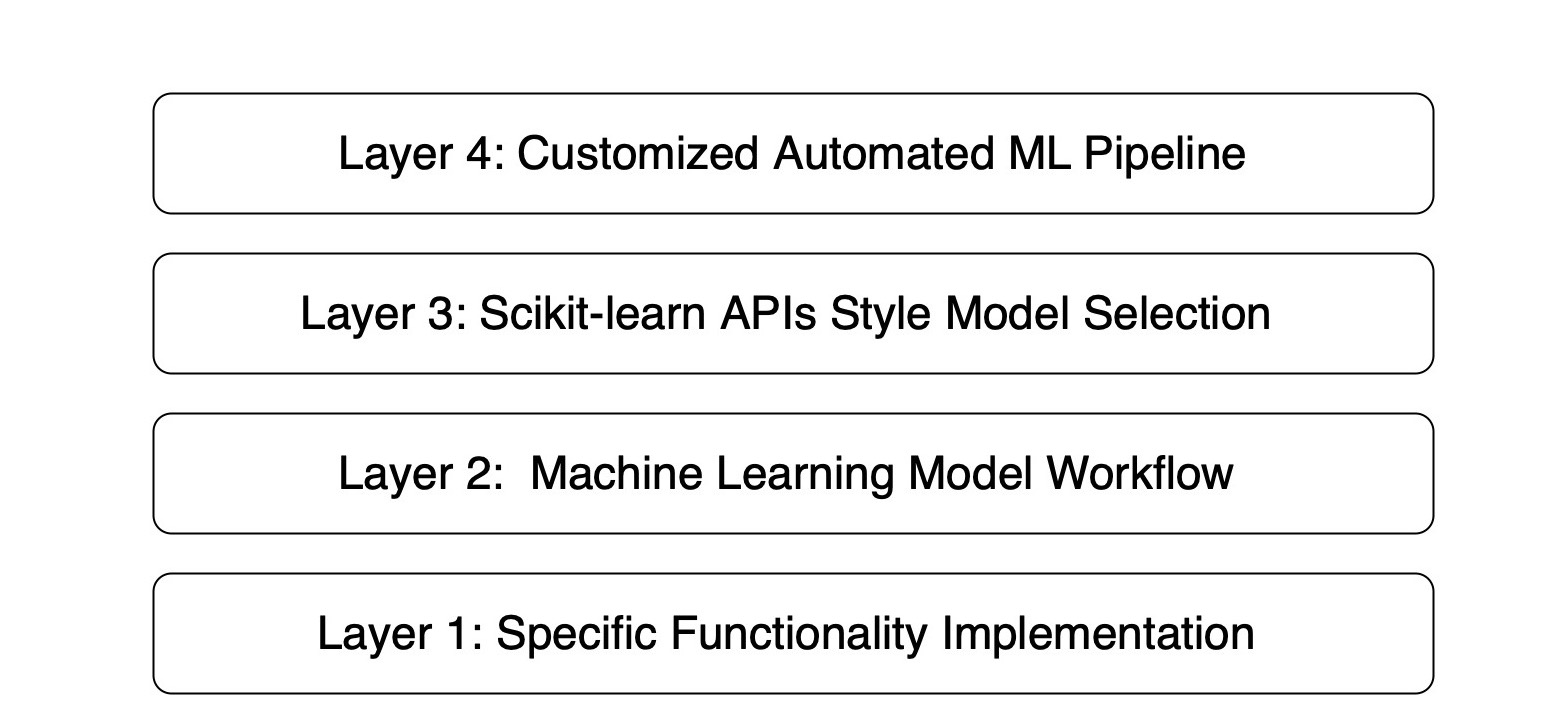
+The following figure is the hierarchical architecture of Geochemistry π:
+
+  +
+
+
### Second Phase
Currently, we are building three access ways to provide more user-friendly service, including **web portal**, **CLI package** and **API**. It allows the user to perform **continuous training** and **model inference** by automating the ML pipeline and **machine learning lifecycle management** by unique storage mechanism in different access layers.
@@ -151,9 +158,9 @@ The following figure is the system architecture diagram:
The following figure is the customized automated ML pipeline:
-
+
 -
-
@@ -162,9 +169,9 @@ The following figure is the design pattern hierarchical architecture:
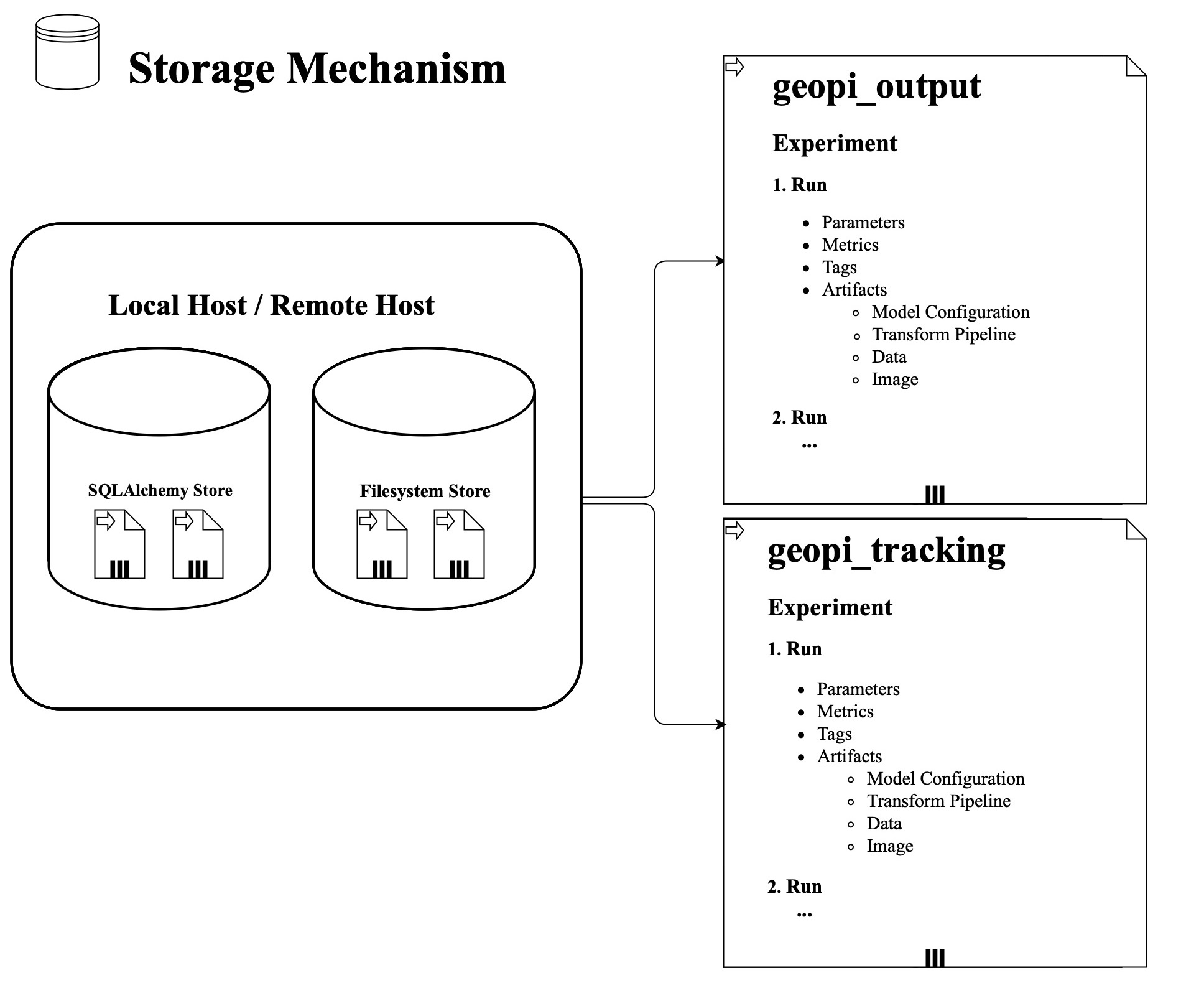
The following figure is the storage mechanism:
-
+
 -
-
+
+
+
-
-