forked from b-rodrigues/chronicler
-
Notifications
You must be signed in to change notification settings - Fork 0
/
Copy pathREADME.Rmd
352 lines (247 loc) · 9.28 KB
/
README.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
---
output: github_document
---
<!-- README.md is generated from README.Rmd. Please edit that file -->
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
```
# chronicler <img src="man/figures/hex.png" align="right" style="width: 25%;"/>
<!-- badges: start -->
<!-- badges: end -->
Easily add logs to your functions, without interfering with the global environment.
## Installation
The package is available on [CRAN](https://cran.r-project.org/web/packages/chronicler/).
Install it with:
```{r, eval = FALSE}
install.packages("chronicler")
```
You can install the development version from [GitHub](https://github.com/) with:
```{r, eval = FALSE}
# install.packages("devtools")
devtools::install_github("b-rodrigues/chronicler")
```
## Introduction
`{chronicler}` provides the `record()` function, which allows you to modify functions so that they
provide enhanced output. This enhanced output consists in a detailed log, and by chaining decorated
functions, it becomes possible to have a complete trace of the operations that led to the final
output. These decorated functions work exactly the same as their undecorated counterparts, but some
care is required for correctly handling them. This introduction will give you a quick overview of
this package’s functionality.
Let's first start with a simple example, by decorating the `sqrt()` function:
```{r example}
library(chronicler)
r_sqrt <- record(sqrt)
a <- r_sqrt(1:5)
```
Object `a` is now an object of class `chronicle`. Let's take a closer look at `a`:
```{r}
a
```
`a` is now made up of several parts. The first part:
```
OK! Value computed successfully:
---------------
Just
[1] 1.000000 1.414214 1.732051 2.000000 2.236068
```
simply provides the result of `sqrt()` applied to `1:5` (let's ignore the word `Just` on the third
line for now; for more details see the `Maybe Monad` vignette). The second part tells you that
there's more to it:
```
---------------
This is an object of type `chronicle`.
Retrieve the value of this object with pick(.c, "value").
To read the log of this object, call read_log().
```
The value of the `sqrt()` function applied to its arguments can be obtained using `pick()`, as
explained:
```{r}
pick(a, "value")
```
A log also gets generated and can be read using `read_log()`:
```{r}
read_log(a)
```
This is especially useful for objects that get created using multiple calls:
```{r}
r_sqrt <- record(sqrt)
r_exp <- record(exp)
r_mean <- record(mean)
b <- 1:10 |>
r_sqrt() |>
bind_record(r_exp) |>
bind_record(r_mean)
```
(`bind_record()` is used to chain multiple decorated functions and will be explained in
detail in the next section.)
```{r}
read_log(b)
pick(b, "value")
```
`record()` works with any function, but not yet with `{ggplot2}`.
To avoid having to define every function individually, like this:
```{r}
r_sqrt <- record(sqrt)
r_exp <- record(exp)
r_mean <- record(mean)
```
you can use the `record_many()` function. `record_many()` takes a list of functions (as strings)
as an input and puts generated code in your system's clipboard. You can then paste the code
into your text editor. The gif below illustrates how `record_many()` works:
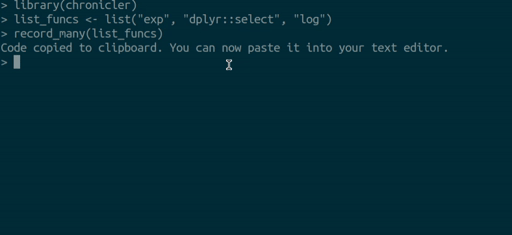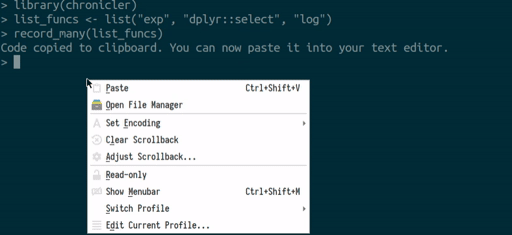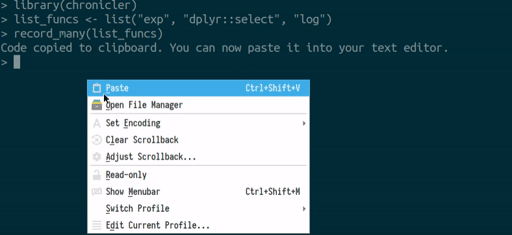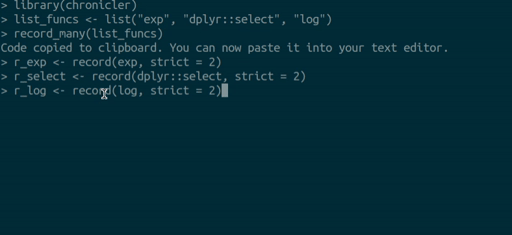
## Chaining decorated functions
`bind_record()` is used to pass the output from one decorated function to the next:
```{r}
library(dplyr)
r_group_by <- record(group_by)
r_select <- record(select)
r_summarise <- record(summarise)
r_filter <- record(filter)
output <- starwars %>%
r_select(height, mass, species, sex) %>%
bind_record(r_group_by, species, sex) %>%
bind_record(r_filter, sex != "male") %>%
bind_record(r_summarise,
mass = mean(mass, na.rm = TRUE)
)
```
```{r}
read_log(output)
```
The value can then be accessed and worked on as usual using `pick()`, as explained above:
```{r}
pick(output, "value")
```
This package also ships with a dedicated pipe, `%>=%` which you can use instead of `bind_record()`:
```{r}
output_pipe <- starwars %>%
r_select(height, mass, species, sex) %>=%
r_group_by(species, sex) %>=%
r_filter(sex != "male") %>=%
r_summarise(mean_mass = mean(mass, na.rm = TRUE))
```
```{r}
pick(output_pipe, "value")
```
Using the `%>=%` is not recommended in non-interactive sessions and `bind_record()`
is recommend in such settings.
## Condition handling
By default, errors and warnings get caught and composed in the log:
```{r}
errord_output <- starwars %>%
r_select(height, mass, species, sex) %>=%
r_group_by(species, sx) %>=% # typo, "sx" instead of "sex"
r_filter(sex != "male") %>=%
r_summarise(mass = mean(mass, na.rm = TRUE))
```
```{r}
errord_output
```
Reading the log tells you which function failed, and with which error message:
```{r}
read_log(errord_output)
```
It is also possible to only capture errors, or capture errors, warnings and messages using
the `strict` parameter of `record()`
```{r}
# Only errors:
r_sqrt <- record(sqrt, strict = 1)
r_sqrt(-10) |>
read_log()
# Errors and warnings:
r_sqrt <- record(sqrt, strict = 2)
r_sqrt(-10) |>
read_log()
# Errors, warnings and messages
my_f <- function(x){
message("this is a message")
10
}
record(my_f, strict = 3)(10) |>
read_log()
```
## Advanced logging
You can provide a function to `record()`, which will be evaluated on the output. This makes it possible
to, for example, monitor the size of a data frame throughout the pipeline:
```{r}
r_group_by <- record(group_by)
r_select <- record(select, .g = dim)
r_summarise <- record(summarise, .g = dim)
r_filter <- record(filter, .g = dim)
output_pipe <- starwars %>%
r_select(height, mass, species, sex) %>=%
r_group_by(species, sex) %>=%
r_filter(sex != "male") %>=%
r_summarise(mass = mean(mass, na.rm = TRUE))
```
The `$log_df` element of a `chronicle` object contains detailed information:
```{r}
pick(output_pipe, "log_df")
```
It is thus possible to take a look at the output of the function provided (`dim()`) using
`check_g()`:
```{r}
check_g(output_pipe)
```
We can see that the dimension of the dataframe was (87, 4) after the call to `select()`, (23, 4)
after the call to `filter()` and finally (9, 3) after the call to `summarise()`.
Another possibility for advanced logging is to use the `diff` argument in record, which defaults
to "none". Setting it to "full" provides, at each step of a workflow, the diff between the input
and the output:
```{r}
r_group_by <- record(group_by)
r_select <- record(select, diff = "full")
r_summarise <- record(summarise, diff = "full")
r_filter <- record(filter, diff = "full")
output_pipe <- starwars %>%
r_select(height, mass, species, sex) %>=%
r_group_by(species, sex) %>=%
r_filter(sex != "male") %>=%
r_summarise(mass = mean(mass, na.rm = TRUE))
```
Let's compare the input and the output to `r_filter(sex != "male")`:
```{r}
# The following line generates a data frame with columns `ops_number`, `function` and `diff_obj`
# it is possible to filter on the step of interest using the `ops_number` or the `function` column
diff_pipe <- check_diff(output_pipe)
diff_pipe %>%
filter(`function` == "filter") %>% # <- backticks around `function` are required
pull(diff_obj)
```
If you are familiar with the version control software `Git`, you should have no problem reading
this output. The input was a data frame of 87 rows and 4 columns, and the output only had 23 rows.
Rows that were in the input, and got removed from the output, are highlighted (in the terminal,
but not here, due to the color scheme).
If `diff` is set to "summary", then only a summary is provided:
```{r}
r_group_by <- record(group_by)
r_select <- record(select, diff = "summary")
r_summarise <- record(summarise, diff = "summary")
r_filter <- record(filter, diff = "summary")
output_pipe <- starwars %>%
r_select(height, mass, species, sex) %>=%
r_group_by(species, sex) %>=%
r_filter(sex != "male") %>=%
r_summarise(mass = mean(mass, na.rm = TRUE))
diff_pipe <- check_diff(output_pipe)
diff_pipe %>%
filter(`function` == "filter") %>% # <- backticks around `function` are required
pull(diff_obj)
```
By combining `.g` and `diff`, it is possible to have a very clear overview of what happened to the very
first input throughout the pipeline.
`diff` functionality is provided by the `{diffobj}` package.
## Thanks
I’d like to thank [armcn](https://github.com/armcn), [Kupac](https://github.com/Kupac) for their
blog posts ([here](https://kupac.gitlab.io/biofunctor/2019/05/25/maybe-monad-in-r/)) and
packages ([maybe](https://armcn.github.io/maybe/)) which inspired me to build this package.
Thank you as well to [TimTeaFan](https://community.rstudio.com/t/help-with-writing-a-custom-pipe-and-environments/133447/2?u=brodriguesco)
for his help with writing the `%>=%` infix operator, [nigrahamuk](https://community.rstudio.com/t/best-way-to-catch-rlang-errors-consistently/131632/5?u=brodriguesco)
for showing me a nice way to catch errors, and finally [Mwavu](https://community.rstudio.com/t/how-to-do-call-a-dplyr-function/131396/2?u=brodriguesco)
for pointing me towards the right direction with an issue I've had as I started working on this package.
Thanks to [Putosaure](https://twitter.com/putosaure) for designing the hex logo.