forked from murphyqm/key-data-vis-requirements
-
Notifications
You must be signed in to change notification settings - Fork 1
/
Copy pathEncoding.py
577 lines (420 loc) · 29.8 KB
/
Encoding.py
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
import streamlit as st
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import numpy as np
import pandas as pd
from matplotlib import colormaps
import matplotlib as mpl
import requests
import scipy as sp
import urllib.request
from PIL import Image
# from bokeh.plotting import figure
#from bokeh.transform import linear_cmap, factor_cmap
#from bokeh.palettes import Bokeh
# page urls
url_home = "https://data-vis-101.streamlit.app/"
url_audience = "https://key-data-vis-requirements-audience.streamlit.app/"
url_story = "https://key-data-vis-requirements-story.streamlit.app/"
url_encoding = "https://key-data-vis-requirements-encoding.streamlit.app/"
url_composition = "https://key-data-vis-requirements-composition.streamlit.app/"
url_simplify = "https://key-data-vis-requirements-simplify.streamlit.app/"
# next_page_url
url_next = url_composition
urls = [url_home, url_audience, url_story, url_encoding, url_composition, url_simplify]
home_title = " Home"
audience_title = "1. Audience"
story_title = "2. Story"
encoding_title = "3. Encoding"
composition_title = "4. Composition"
simplify_title = "5. Simplify"
titles = [home_title, audience_title , story_title, encoding_title, composition_title, simplify_title]
def click_button(url, text):
st.markdown(f'<a style="word-wrap:break-word;color:#ea388f;font-size:20px;font-style:bold; width: fit-content; border-style: none;padding:6px;text-decoration: none;border-width:2px;" href={url} target="_self">{text}</a>', unsafe_allow_html=True)
def pink_button(url, text):
st.markdown(f'<a style="word-wrap:break-word;color:#ea388f;font-size:20px;font-style:bold; width: fit-content; border-style: solid;border-radius:10px;padding:6px;text-decoration: none;border-width:2px;text-align: center;" href={url} target="_self">{text}</a>', unsafe_allow_html=True)
def nav_bar(urls, titles):
with st.popover("Table of contents", use_container_width=True):
click_button(urls[0], titles[0])
click_button(urls[1], titles[1])
click_button(urls[2], titles[2])
click_button(urls[3], titles[3])
click_button(urls[4], titles[4])
click_button(urls[5], titles[5])
def next_page(url_next):
pink_button(url_next, "Next section →")
@st.cache_data
def load_image_from_github(url):
img = np.array(Image.open(requests.get(url, stream=True).raw))
return img
my_pall ={'orange': '#e66101',
'pink': '#CA054D',
'blue': '#1B98E0',
'green':'#A4D4B4',
'purple': '#5e3c99', }
# data analysis
penguins = sns.load_dataset("penguins").dropna()
Penguins = penguins.rename(columns={"species": "Species",
"island": "Island",
"bill_length_mm": "Bill length (mm)",
"bill_depth_mm": "Bill depth (mm)",
"flipper_length_mm": "Flipper length (mm)",
"body_mass_g": "Body mass (g)",
"sex":"Sex"
})
# colour maps
# peng_cmap = factor_cmap(penguins, palette=Bokeh, factors=sorted(penguins.species.unique()))
# building data for ternary plot
locat = ["Locality #01B-32", "Locality #01B-34", "Locality #02C-01"]
local_no = np.random.choice(locat, 200)
local_var = np.zeros(200)
local_var [local_no == "Locality #01B-32"] = 1.3
local_var [local_no == "Locality #01B-34"] = -1.2
Qm_raw = 3.5 * np.random.randn(200) + 5
Qm_raw = Qm_raw + local_var
Qm_raw [Qm_raw <0] = 1.3
F_raw = 1.5 * np.random.randn(200) + 3
F_raw [F_raw <0] = 0.5
Lt_raw = 0.7 * np.random.randn(200) + 2
Lt_raw = Lt_raw - local_var
Lt_raw[Lt_raw <0] = 0.1
total_val = Qm_raw + F_raw + Lt_raw
norm_factor = 100/total_val
Qm = Qm_raw * norm_factor
F = F_raw * norm_factor
Lt = Lt_raw * norm_factor
geo_data = {"Qm":Qm,
"F":F,
"Lt":Lt,
"Locality Number": local_no}
geo_df = pd.DataFrame.from_dict(geo_data)
st.title('5 Concepts for Data Visualisation')
intro_text = """
This website introduces five key concepts to help you build better research data visualisations.
These are not the *only* rules, or the *only* way to approach building a robust and reproducible
graphical research output, but should help you to create visualisations in a methodical way.
"""
st.write(intro_text)
col1, col2 = st.columns(2)
pub_refs = """
# Sources and inspiration
A number of fantastic articles went into this resource. Please read these works: the content shared here is a much-abridged, simplified, shortened version of the content shared in the articles below.
#### Bibliography
Berinato, Scott. 2016. “Visualizations That Really Work.” Harvard Business Review, June 1, 2016. https://hbr.org/2016/06/visualizations-that-really-work.
Franconeri, Steven L., Lace M. Padilla, Priti Shah, Jeffrey M. Zacks, and Jessica Hullman. 2021. “The Science of Visual Data Communication: What Works.” Psychological Science in the Public Interest: A Journal of the American Psychological Society 22 (3): 110–61.
Kelleher, Christa, and Thorsten Wagener. 2011. “Ten Guidelines for Effective Data Visualization in Scientific Publications.” Environmental Modelling & Software: With Environment Data News 26 (6): 822–27.
Midway, Stephen R. 2020. “Principles of Effective Data Visualization.” Patterns (New York, N.Y.) 1 (9): 100141.
Rougier, Nicolas P., Michael Droettboom, and Philip E. Bourne. 2014. “Ten Simple Rules for Better Figures.” PLoS Computational Biology 10 (9): e1003833.
#### Other resources
- [Berkeley Library Data Visualisation library guide](https://guides.lib.berkeley.edu/data-visualization/about)
- [Tableau documentation: Visual Best Practices](https://help.tableau.com/current/blueprint/en-us/bp_visual_best_practices.htm)
- [from Data to Viz project](https://www.data-to-viz.com/)
- [Principles of Data Visualization workshop notes](https://ucdavisdatalab.github.io/workshop_data_viz_principles/)
- [What’s visual ‘encoding’ in data viz, and why is it important?](https://medium.com/@sophiewarnes/whats-visual-encoding-in-data-viz-and-why-is-it-important-7406bc88b4b4#:~:text=Encoding%20in%20data%20viz%20basically,trying%20to%20say%20or%20show.)
- [University of Utah visualization design lab](https://vdl.sci.utah.edu/)
"""
with col1:
with st.popover("Publications and references behind this tool", use_container_width=True):
st.markdown(pub_refs)
package_references = """
#### matplotlib

J. D. Hunter, "Matplotlib: A 2D Graphics Environment", Computing in Science & Engineering, vol. 9, no. 3, pp. 90-95, 2007.
#### seaborn

Waskom, M. L., (2021). seaborn: statistical data visualization. Journal of Open Source Software, 6(60), 3021, https://doi.org/10.21105/joss.03021.
#### Plotly

Plotly Technologies Inc. Collaborative data science. Montréal, QC, 2015. https://plot.ly.
#### Streamlit

Streamlit documentation, https://docs.streamlit.io/
#### NumPy

Harris, C.R., Millman, K.J., van der Walt, S.J. et al. Array programming with NumPy. Nature 585, 357–362 (2020). DOI: 10.1038/s41586-020-2649-2.
### pandas

Data structures for statistical computing in python, McKinney, Proceedings of the 9th Python in Science Conference, Volume 445, 2010.
"""
with col2:
with st.popover("Python packages and datasets used", use_container_width=True):
st.header("Python packages")
st.markdown(package_references)
st.header("Datasets")
st.write("We use the `penguins` example dataset from the seaborn library. This can be browsed below:")
st.dataframe(penguins)
st.write("We also generated some random geological data to build a QFL ternary plot. This will regenerate when you reload the page, but will look something similar to this:")
st.dataframe(geo_df)
# # nav_bar_2(urls, titles)
# # st.markdown("", unsafe_allow_html=False)
# nav_bar(urls, titles)
encoding_01 = """
When discussing data visualisation, **data encoding** essentially means *how you translate the data into a visual element on a chart* ([Warnes, 2018](https://medium.com/@sophiewarnes/whats-visual-encoding-in-data-viz-and-why-is-it-important-7406bc88b4b4#:~:text=Encoding%20in%20data%20viz%20basically,trying%20to%20say%20or%20show.)). In her fantastic blog post, [Sophie Warnes](https://medium.com/@sophiewarnes/whats-visual-encoding-in-data-viz-and-why-is-it-important-7406bc88b4b4#:~:text=Encoding%20in%20data%20viz%20basically,trying%20to%20say%20or%20show.) describes data encoding as a set of rules to follow, giving the following logic:
> *Every time <data changes in some way>, do <something visual>,*
where the *something visual* is the encoding.
Starting simply, lets consider a scatter plot. We can think about how we might change a point marker to encode the data. There are many different ways we can encode data in this setting, including but not limited to:
- **Position**
- The *x* and *y* co-ordinates reflect the data
- **Colour**
- Colour can be used to represent a continuous numerical value (colour map)
- Colour can be used to represent discrete or categorical data
- **Shape**
- Marker shape could be used to represent categorical data or highlight specific points
- **Size**
- Size could be used to represent a continuous numerical value or category
> How many distinct values do you need to discriminate between? This can define the method of encoding that best suits your needs.
We're going to look at two main factors as we discuss encoding data:
##### 1. Truthful and non-misleading graphics
##### 2. Easy to read, efficient graphics
---
Take some time to read through the UCDavis DataLab [Principles of Visual Perception](https://ucdavisdatalab.github.io/workshop_data_viz_principles/principles-of-visual-perception.html) section in their data visualisation workshop notes.
They highlight a number of ways we can be misled by graphics, and how we can leverage the way our brains absorb visual information to build better data visualisations. For example, see the figure below:
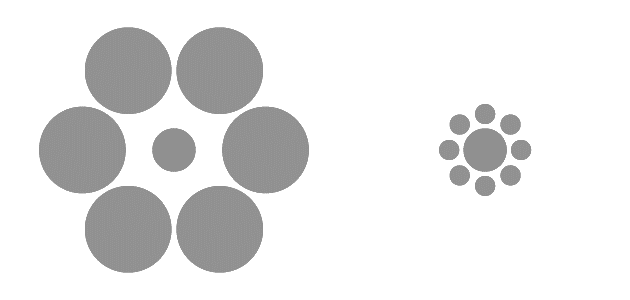
*Caption: Which inner circle is bigger? From [UCDavis Principles of data Visualisation workshop](https://ucdavisdatalab.github.io/workshop_data_viz_principles/principles-of-visual-perception.html#visual-magic-tricks)*
---
## 1. Building truthful and non-misleading graphics
> How effective are the different methods of encoding data shown below? What are some limitations or drawbacks of each option?
"""
encoding_02 = """
## 2. Building easy to read, efficient graphics
We want people to be able to quickly absorb information from our plots. Research generally handles complex topics, and the visualisations that we build are often trying to distil the results of months or years of complex work. We want our audience to simultaneously scan our figure, read labels and annotations, and absorb the salient points. This is a lot to juggle, and it's in our best interests to reduce the cognitive load and the strain on the visual working memory of our reader.
Thankfully, there's a whole host of research on how to best create visualisations that allow the audience to quickly absorb information and to leverage intuitive shortcuts to make our plots more digestible.
#### Pre-attentive attributes
The [Tableau documentation](https://help.tableau.com/current/blueprint/en-us/bp_why_visual_analytics.htm) defines pre-attentive attributes as **"information we can process visually almost immediately"**, without thinking.
These attributes include [length, width, orientation, size, shape, enclosure, position, grouping, colour hue, and colour intensity](https://help.tableau.com/current/blueprint/en-us/Img/bp_why_visual_analytics.png). Leveraging these encoding channels can help to make your visualisation more rapidly understandable.
These attributes can be ranked according to the accuracy with which we can estimate their value: position and length are usually accurately estimated by the viewer (with sensible axes labels, scales, placement etc.), followed by angle and slope, then area, volume, colour, and density/saturation ([Mackinlay, 1986](https://dl.acm.org/doi/10.1145/22949.22950); [UCDavis DataLab](https://ucdavisdatalab.github.io/workshop_data_viz_principles/principles-of-visual-perception.html#perception-and-encodings)).
#### Gestalt principles of grouping
As well as looking at the effect of the individual encoding of datapoints, we need to consider the plot as a whole. Gestalt psychology looks at how people perceive things as a whole entity rather than as separate parts. Without the time to dive into the backgroiund of this field of psychology and address some of the assumptions and caveats (see [Table 2, Wagemans et al., 2012](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3482144/)), lets briefly look at some of the principles of grouping that can be applied to your figure making.
- **Proximity**
- How near to each other objects are
- Our brain performs automatic clustering of points, and considers items close to each other as related
- This can be used...
- When arranging plots in a facet layout or a grid
- When arranging 1D plots such as bar charts
- When creating visualisations without numeric values on the *x* or *y* axes
- **Similarity**
- How similar objects are to each other
- Using the encoding channels explained above for marker style to group points
- Use [this graphic](https://www.oreilly.com/library/view/designing-data-visualizations/9781449314774/ch04.html#use_this_table_of_common_visual_properti) to pick an appropriate channel for the number of discrete values/groups you need to identify
- **Connection**
- Whether data points are isolated or connected
- Points that are connected to each other (via a line) are perceived as related
- Line style, colour and opacity can be varied
- **Enclosure**
- Whether points are encircled in a group or have a different background colour/shading in a region
- Points enclosed in a region are perceived as related
- Annotations can be added to enclosed regions
For further information, please see [UCDavis DataLab Principles of Data Visualisation notes](https://ucdavisdatalab.github.io/workshop_data_viz_principles/principles-of-visual-perception.html#gestalt-principles), [Wagemans et al., 2012](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3482144/), and [Peterson and Berryhill, 2013](https://link.springer.com/article/10.3758/s13423-013-0460-x)
"""
encoding_refs = """
**References**
Berinato, Scott. 2016. “Visualizations That Really Work.” Harvard Business Review, June 1, 2016. https://hbr.org/2016/06/visualizations-that-really-work.
Franconeri, Steven L., Lace M. Padilla, Priti Shah, Jeffrey M. Zacks, and Jessica Hullman. 2021. “The Science of Visual Data Communication: What Works.” Psychological Science in the Public Interest: A Journal of the American Psychological Society 22 (3): 110–61.
Kelleher, Christa, and Thorsten Wagener. 2011. “Ten Guidelines for Effective Data Visualization in Scientific Publications.” Environmental Modelling & Software: With Environment Data News 26 (6): 822–27.
Wagemans, Johan, James H. Elder, Michael Kubovy, Stephen E. Palmer, Mary A. Peterson, Manish Singh, and Rüdiger von der Heydt. 2012. “A Century of Gestalt Psychology in Visual Perception: I. Perceptual Grouping and Figure-Ground Organization.” Psychological Bulletin 138 (6): 1172–1217.
"""
colour_choice = """
Some things to keep in mind when picking a colour palette for your figure:
1. Is the data ordered or not? Do you need to use a **qualitative**, **sequential** or **diverging** palette?
2. How many levels need to be distinguishable?
3. Will the plot be readable if you have a colour vision deficiency (i.e., colour blindness)?
4. Will the plot be readable if printed in black and white?
5. Is there enough contrast between the different colours that it will be legible on a range of different screens?
You can avail of tools such as [ColorBrewer](https://colorbrewer2.org/) to pick a basic scientific palette. For more fine-grained control, try using [Chroma.js](https://gka.github.io/palettes/#/9|s|00429d,96ffea,ffffe0|ffffe0,ff005e,93003a|1|1) or [Colorgorical](http://vrl.cs.brown.edu/color) to generate palettes.
If you already have a palette in mind, test it out with the fantastic [Viz Palette](https://projects.susielu.com/viz-palette) tool. This will allow you to export hex codes of colours to use in Python.
Further reading:
- [Your Friendly Guide to Colors in Data Visualisation](https://blog.datawrapper.de/colorguide/)
- [Best Color Palettes for Scientific Figures and Data Visualizations](https://www.simplifiedsciencepublishing.com/resources/best-color-palettes-for-scientific-figures-and-data-visualizations#:~:text=How%20to%20Find%20the%20Best,and%20other%20color%20perception%20difficulties.)
"""
heatmaps_md = """
There is a wealth of information and research into choosing the best heatmap to illustrate your data.
[Crameri et al. (2020)](https://www.nature.com/articles/s41467-020-19160-7) highlight some of the issues with using poorly designed heatmaps:
> Zones of danger, such as the boundaries of a hurricane track or current virus spread, are often based on uneven colour gradients to accentuate their importance. [...] Decisions based on data being ‘unfairly’ represented could produce, for instance, a Martian rover being sent over terrain that is too steep as the topography was inaccurately visualised, or a medical worker making an incomplete or inaccurate diagnosis based on uneven colour gradients.
- You can see in the examples above how the `spring` heatmap squashes details at the extremes of the data, so you lose information near the max and minimum values.
- While `jet` allows us to see detail in the extremes of the data, it also produces artifacts due to jumps in lightness between colours.
- The `plasma` colourmap removes these jump artifacts, and allows us to see more in the extreme values than `spring` did.
- The `turbo` colourmap has been designed as a replacement for [`jet`](https://research.google/blog/turbo-an-improved-rainbow-colormap-for-visualization/), where "where perceptual uniformity is not critical, but one still wants a high contrast, smooth visualization of the underlying data". This allows maxima and minima to be clearly seen, and doesn't produce artifical large jumps in brightness.
See the [matplotlib documentation on colour maps for examples](https://matplotlib.org/stable/users/explain/colors/colormaps.html#lightness-of-matplotlib-colormaps).
Other heatmap tools for Python include ["batlow": the Scientific colour map](https://www.fabiocrameri.ch/batlow/), [seaborn colour maps](https://seaborn.pydata.org/tutorial/color_palettes.html#sequential-color-palettes), [cmocean colour maps](https://matplotlib.org/cmocean/), [the SciCoMap package](https://github.com/ThomasBury/scicomap) and the related [blog pot](https://towardsdatascience.com/your-colour-map-is-bad-heres-how-to-fix-it-lessons-learnt-from-the-event-horizon-telescope-b82523f09469)
"""
cmaps = {}
gradient = np.linspace(0, 1, 256)
gradient = np.vstack((gradient, gradient))
def plot_color_gradients(category, cmap_list):
# Create figure and adjust figure height to number of colormaps
nrows = len(cmap_list)
figh = 0.35 + 0.15 + (nrows + (nrows - 1) * 0.1) * 0.22
fig, axs = plt.subplots(nrows=nrows + 1, figsize=(6.4, figh))
fig.subplots_adjust(top=1 - 0.35 / figh, bottom=0.15 / figh,
left=0.2, right=0.99)
axs[0].set_title(f'{category} colormaps', fontsize=14)
for ax, name in zip(axs, cmap_list):
ax.imshow(gradient, aspect='auto', cmap=mpl.colormaps[name])
ax.text(-0.01, 0.5, name, va='center', ha='right', fontsize=10,
transform=ax.transAxes)
# Turn off *all* ticks & spines, not just the ones with colormaps.
for ax in axs:
ax.set_axis_off()
# Save colormap list for later.
cmaps[category] = cmap_list
return fig
st.title("3. Encoding")
st.markdown(encoding_01)
with st.expander("Compare different encoding methods"):
if "encoding" not in st.session_state:
st.session_state.encoding = "All the above"
st.radio("Pick an *encoding channel*:", key="encoding",
options=["Marker shape", "Marker colour", "Marker size", "All the above"])
args_full = {"style":"Island", "hue":"Island", "size":"Island"}
args_style = {"style":"Island"}
args_hue = {"hue":"Island"}
args_size = {"size":"Island"}
if st.session_state.encoding == "All the above":
args_ = args_full
elif st.session_state.encoding == "Marker shape":
args_ = args_style
elif st.session_state.encoding == "Marker colour":
args_ = args_hue
elif st.session_state.encoding == "Marker size":
args_ = args_size
else:
args_ = args_full
fig7 = plt.figure()
with sns.axes_style("ticks"):
sns.set_context("talk")
sns.set_palette("Set2")
ax = sns.scatterplot(data=Penguins, x="Body mass (g)", y="Flipper length (mm)", alpha=0.9, **args_ )
sns.despine()
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1), title="Encoding")
st.pyplot(fig7, use_container_width=True,)
st.write("Refer to [this useful graphic](https://www.oreilly.com/library/view/designing-data-visualizations/9781449314774/ch04.html#use_this_table_of_common_visual_properti) when trying to decide what encoding channel to use.")
with st.expander("Size of marker as an encoding channel..."):
st.write("Size is an attribute that is easy to set to map onto a numerical value, to provide a third dimension to your visualisation.",
"However, there are a few issues with this approach.")
st.subheader("1. We are not very good at estimating area")
st.write("It's difficult to accurately estimate the area of a marker, making it difficult for readers to decode our visualisation. This is also why it's best to [avoid pie-charts wherever possible](https://theconversation.com/heres-why-you-should-almost-never-use-a-pie-chart-for-your-data-214576#:~:text=Pie%20charts%20also%20do%20badly,of%20categories%20in%20one%20pie.&text=The%20tiny%20slices%2C%20lack%20of,make%20interpretation%20difficult%20for%20anyone.)")
st.subheader("2. It's not always clear or obvious the component of 'size' that maps onto the value, or how it scales")
st.write("Different Python plotting libraries tie different marker attributes to the idea of 'size' - marker diameter, radius, and area. This means that different marker shapes masy scale differently for different packages.")
st.write("Then depending on the scale of your data, you may have to use a multiple or factor to adjust the size relationship to make it visible in your plot. See below for the different effects this can have.")
x = [0,2,4,6,8,10,12,14,16,18]
s_exp = [20*2**n for n in range(len(x))]
s_square = [20*n**2 for n in range(len(x))]
s_linear = [20*n for n in range(len(x))]
fig_05, ax = plt.subplots()
ax.scatter(x,[1]*len(x),s=s_exp, label='$s=20 * 2^x$', lw=1, alpha=0.6, c=my_pall["pink"])
ax.scatter(x,[0]*len(x),s=s_square, label='$s=20 * x^2$', alpha=0.7, c=my_pall["blue"])
ax.scatter(x,[-1]*len(x),s=s_linear, label='$s=20 * x$', alpha=0.7, c=my_pall["orange"])
for i in x:
ax.text(i, -1.6, str(i), horizontalalignment='center', verticalalignment='center')
ax.axes.vlines(x=22, ymin=-1.7, ymax=1.6, colors="grey", linestyles=":", linewidths=2)
ax.set_ylim(-2,2)
ax.set_xlim(-0.5, 22)
ax.set_xlabel("X")
ax.legend(loc='center left', bbox_to_anchor=(1.05, 0.45), labelspacing=6, frameon=False, handletextpad=3.5)
ax.spines[:].set_visible(False)
ax.set_yticks([])
ax.set_xticks([])
st.pyplot(fig_05, use_container_width=True,)
st.write("*Caption: Size (s) in matplotlib library set as different mutliples of x value.*")
st.subheader("Implying order")
with st.expander("How might data encoding imply that the data is ordered?"):
st.write("Both these figures use the same randomly generated data.",
"What inferences might you draw from each of these plots? How is this influenced by the encoding of the data?")
# Create a figure and axes, and set the figure size in inches
fig8, ax = plt.subplots(figsize=(7, 4))
# Random data
x = np.random.randn(50) + 2
y = np.random.randn(50) - 1
x1 = np.random.randn(50)
y1 = np.random.randn(50)
x2 = np.random.randn(50) - 2
y2 = np.random.randn(50) - 1
x3 = np.random.randn(50) + 2
y3 = np.random.randn(50) + 2
# Plot the data as a scatter plot
ax.scatter(x, y, label="Group A", c=my_pall["pink"], alpha=0.5, marker="*", s=130)
ax.scatter(x1, y1, label="Group B", c=my_pall["blue"], alpha=0.5, marker="X", s=130)
ax.scatter(x2, y2, label = "Group C", c=my_pall["orange"], alpha=0.5, s=120)
ax.scatter(x3, y3, label = "Group D", c=my_pall["purple"], alpha=0.5, s=110, marker="s")
# Set title and labels
ax.legend()
ax.set_title('Scatter Plot')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.spines[["top", "right"]].set_visible(False)
st.pyplot(fig8, use_container_width=True,)
fig9, ax = plt.subplots(figsize=(7, 4))
new_pall = ["#cbc9e2",
"#9e9ac8",
"#756bb1",
"#54278f",]
# Plot the data as a scatter plot
ax.scatter(x, y, label="Group A", c=new_pall[0], alpha=0.8, s=200)
ax.scatter(x1, y1, label="Group B", c=new_pall[1], alpha=0.8, s=150)
ax.scatter(x2, y2, label = "Group C", c=new_pall[2], alpha=0.8, s=100)
ax.scatter(x3, y3, label = "Group D", c=new_pall[3], alpha=0.8, s=50, )
# Set title and labels
ax.legend()
ax.set_title('Scatter Plot')
ax.set_xlabel('X')
ax.set_ylabel('Y')
ax.spines[["top", "right"]].set_visible(False)
st.pyplot(fig9, use_container_width=True,)
st.write("*Caption: Randomly generated data using different shape, size and colour encoding.*")
st.write("Order can be implied by colour, size, position, angle (of marker). Ensure that you do not unintentionally imply that data are ordered when they are not.",
"When listing data sets in the legend, if there is no scientific basis for ordering them in a specific way, alphabetise them, so as not to unintentionally apply a ranking. For example, country names should be alphabetised inside alphabetised continent headers.")
with st.expander("Colour always means something"):
st.write("Be mindful when selecting colours for your plots.",
"It's good to keep in mind what colour might represent to your audience, and if this is what you intended.",
"For example, when building a plot discussing political party results,",
"the colours red and blue will have different meanings and interpretation depending on the country.",
"Similarly, using a continuous colour map, or varying saturation and intensity can imply that the data is ordered, as in the above example.")
st.markdown(colour_choice)
st.subheader("Implying contrast")
st.write("Colour, markersize, location, opacity/saturation and labels can be used to emphasise contrast. Later in the course, we will also discuss how axes scaling can be used to artifically increase contrast between datapoints.")
st.write("Unintentional emphasis on contrast can most easily creep in when using continuous colour maps to illustrate data.")
with st.popover("Compare different colour maps (N.B. this will stay open as you scroll)"):
if "heatmap" not in st.session_state:
st.session_state.heatmap = "plasma"
if "ex_image" not in st.session_state:
st.session_state.ex_image = "https://www.leeds.ac.uk/images/resized/1200x600-0-0-1-80-ParkinsonBuilding_2.jpg"
st.radio("Choose a matplotlib heatmap:",key="heatmap",
options=["plasma","jet","turbo","spring"])
jpg_image =load_image_from_github("https://astropedia.astrogeology.usgs.gov/download/Mars/GlobalSurveyor/MOLA/thumbs/Mars_MGS_MOLA_DEM_mosaic_global_1024.jpg)")
image = jpg_image[:,:,0]
image = ((21229+8200)*(image/255)) - 8200
fig, ax = plt.subplots(layout='constrained')
with sns.axes_style("ticks"):
sns.set_context("paper")
im = ax.imshow(image, cmap=st.session_state.heatmap, extent=[-180, 180, -90, 90])
fig.colorbar(im, orientation="horizontal", label="Elevation relative to areoid [M]")
ax.set_xlabel("Longitude")
ax.set_ylabel("Latitude")
st.pyplot(fig, use_container_width=True,)
st.write("Sometimes this is more easily seen with a photograph. Enter your photograph url below.")
ex_im = st.text_input("image URL", "https://www.leeds.ac.uk/images/resized/1200x600-0-0-1-80-ParkinsonBuilding_2.jpg")
new_im = load_image_from_github(str(ex_im))
image = new_im[:,:,0]
image = ((21229+8200)*(image/255)) - 8200
fig_new, ax = plt.subplots(layout='constrained')
with sns.axes_style("white"):
sns.set_context("paper")
im = ax.imshow(image, cmap=st.session_state.heatmap)
ax.set(xticklabels=[], yticklabels=[])
ax.tick_params(left=False,bottom=False)
sns.despine(left=True, bottom=True)
st.pyplot(fig_new, use_container_width=True,)
with st.expander("Picking heatmaps"):
st.write(heatmaps_md)
st.markdown(encoding_02)
st.divider()
st.markdown(encoding_refs)
# st.text("")
# st.text("")
# pink_button(url_next, "Next section →")
st.text("")
st.text("")
st.divider()
st.write(r"$\textsf{\scriptsize Authored by Dr Maeve Murphy Quinlan, University of Leeds Research Computing Team © Copyright 2024}$")
st.write("[App home page](https://data-vis-101.streamlit.app/) | [Research Computing Team](https://arc.leeds.ac.uk/about/team/) | [Research Computing Website](https://arc.leeds.ac.uk/)")